LSTM timeseries forecasting with Keras Tuner
A example of using an LSTM network to forecast timeseries, using Keras Tuner for hyperparameters tuning.
About
This project is a demonstration of some of capabilities of Keras Tuner. This project is an attempt to use an LSTM based neural network (RNN) to forecast timeseries data.

Import the must-have libraries:
import numpy as np
import pandas as pd
import datetime as dt
from IPython import display as ids
Import the elements required from the scikit-learn library:
import sklearn as sk
import sklearn.preprocessing as skp
import sklearn.model_selection as skms
import sklearn.pipeline as skpl
import sklearn.decomposition as skd
import sklearn.linear_model as sklm
import sklearn.dummy as sky
import sklearn.metrics as skme
Enables defining partial functions:
from functools import partial
Import the keras elements from the tensorflow library:
import tensorflow as tf
from tensorflow import keras as k
from tensorflow.keras import backend as kb
from tensorflow.keras import callbacks as kc
from tensorflow.keras import models as km
from tensorflow.keras import layers as kl
from tensorflow.keras import regularizers as kr
from tensorflow.keras import optimizers as ko
from tensorflow.keras import utils as ku
Import the keras-tuner library as we'll use it to tune hyperparameters:
import kerastuner as kt
from kerastuner import tuners as ktt
from kerastuner import engine as kte
Import matplotlib and set the default magic:
import matplotlib as mat
from matplotlib import pyplot as plt
import pylab as pyl
import seaborn as sns
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
Import the mlviz library used to plot time-series visualizations:
from mlviz.timeseries import visualizationhelpers as mwvh
from mlviz.utilities import graphichelpers as mwgh
Reset the predefined matplotlib style:
mwgh.GraphicsStatics.initialize_matplotlib_styles()
Here is the color palette we'll use for the project:
sns.palplot(mwgh.GraphicsStatics.g_palette)
Let's capture all the usefull project's parameters in a dictionary:
params = {}
params['project_date'] = '2021-05-31'
params['project_name'] = 'LSTM+01'
params['experiment_timestamp'] = str(int(1000 * dt.datetime.timestamp(dt.datetime.utcnow())))
params['experiment_name'] = '{0}-{1}-{2}'.format(
params['project_date'],
params['experiment_timestamp'],
params['project_name'])
params['data_frequency'] = 'W-SAT'
params['features'] = ['y']
params['input_size'] = 52 # N weeks of data to input into the network
params['output_size'] = 13 # N weeks of data to output from the network
params['testing_size'] = 52 # N weeks of data to keep for testing the model
params['lag_size'] = [13, 26, 39] # Lagged data series injected into the network
params['hyperband_iterations'] = 3
params['max_epochs'] = 250
params['patience'] = int(params['max_epochs'] / 10)
params['batch_size'] = 32
The input data is available in a csv file named timeseries-data.csv located in the data folder. It has got 2 columns date containing the date of event and value holding the value of the source. We'll rename these 2 columns as ds and y for convenience. Let's load the csv file using the pandas library and have a look at the data.
df = pd.read_csv(
filepath_or_buffer='../assets/data/timeseries-data.csv',
sep=';')
df.rename(
columns = {
'date': 'index',
'value': 'y'
},
inplace=True)
df['index'] = pd.to_datetime(
arg=df['index'],
dayfirst=True)
df.sort_values(
by='index',
ascending=True,
inplace=True)
df.set_index(
keys='index',
inplace=True)
df = df.asfreq(
freq=params['data_frequency'])
df['ds'] = df.index
print('df.shape = {0}'.format(df.shape))
df.tail(5)
It is time to prepare the dataset to feed into the LSTM model. We'll use 4 features as input: the current data point as well as 3 additional data points from respectively 13, 26 and 39 weeks before the current datapoint the lag features below).

It's easy to calculate the width of a sample:
sample_width = max(params['lag_size']) + params['input_size'] + params['output_size']
print('sample_width: {0}'.format(sample_width))
To avoid any overlap between the training and the testing data set, we'll first split the dataframes, keeping params['testing_size'] samples for testing our model. We need to make sure that no data point used for training is also used for testing our model.
threshold_date = pd.to_datetime(df.index[df.shape[0] - (sample_width + params['testing_size'])])
print('Cutoff date for training/testing split is {0}'.format(threshold_date.strftime('%d/%m/%Y')))
Let's cut the dataframe at the right date:
test_mask = (df['ds'] > threshold_date)
df_train = df[~test_mask]
df_test = df[test_mask]
print('df_train.shape = {0}'.format(df_train.shape))
print('df_test.shape = {0}'.format(df_test.shape))
The prepare_data funtion will take care of doing exactly this:
def prepare_data(data, lag_data, cols_in, steps_in, cols_out, steps_out, scaler_in=None, scaler_out=None):
df = data.copy()
cols_in_original = [col for col in cols_in]
cols_in_processed = [col for col in cols_in]
steps_lag = 0
if lag_data is not None and len(lag_data) > 0:
steps_lag = max(lag_data)
for col in cols_in_original:
for i, lag in enumerate(lag_data):
lag_col = '{0}_{1}'.format(col, lag)
df[lag_col] = df[col].shift(lag)
cols_in_processed.append(lag_col)
samples = df.shape[0] - (steps_in + steps_out + steps_lag) + 1
if samples < 1:
raise ValueError('not enough data to produce 1 sample.')
index = list(df.index)
cols_in_indices = {name: i for i, name in enumerate(cols_in_processed)}
cols_out_indices = {name: i for i, name in enumerate(cols_out)}
df.reset_index(inplace=True)
X_input_scaled = None
if scaler_in is None:
scaler_in = skpl.Pipeline([
('std', skp.StandardScaler()),
('minmax', skp.MinMaxScaler(feature_range=(-1, 1)))])
X_input_scaled = scaler_in.fit_transform(df[cols_in_processed].values)
else:
X_input_scaled = scaler_in.transform(df[cols_in_processed].values)
y_output_scaled = None
if scaler_out is None:
scaler_out = skpl.Pipeline([
('std', skp.StandardScaler()),
('minmax', skp.MinMaxScaler(feature_range=(-1, 1)))])
y_output_scaled = scaler_out.fit_transform(df[cols_out].values)
else:
y_output_scaled = scaler_out.transform(df[cols_out].values)
X = []
y = []
for sample in range(samples):
for step_in in range(steps_in):
for col_in in range(len(cols_in_processed)):
X.append(X_input_scaled[sample+steps_lag+step_in, col_in])
for step_out in range(steps_out):
for col_out in range(len(cols_out)):
y.append(y_output_scaled[sample+steps_lag+steps_in+step_out, col_out])
X = np.array(X).reshape(samples, steps_in, len(cols_in_processed))
y = np.array(y).reshape(samples, steps_out, len(cols_out))
return X, y, index, scaler_in, scaler_out, cols_in_indices, cols_out_indices
Every intput feature (passed via the function parameter cols_in) is going to be rescalled using a scikit-learn pipeline containing first a StandardScaler and then a MinMaxScaler in order to end up with a feature range of [-1, +1] required by neural networks.
X_train, y_train, index_train, scaler_in, scaler_out, cols_in_indices_train, cols_out_indices_train = prepare_data(
data=df_train,
lag_data=params['lag_size'],
cols_in=params['features'],
steps_in=params['input_size'],
cols_out=params['features'],
steps_out=params['output_size'])
print('X_train.shape: {0}'.format(X_train.shape))
print('y_train.shape: {0}'.format(y_train.shape))
To prepare the testing data, we need to reuse both input (variable scaler_in) and output (variable scaler_out) pipelines in order to keep data scaled in the same way.
X_test, y_test, index_test, _, _, cols_in_indices_test, cols_out_indices_test = prepare_data(
data=df_test,
lag_data=params['lag_size'],
cols_in=params['features'],
steps_in=params['input_size'],
cols_out=params['features'],
steps_out=params['output_size'],
scaler_in=scaler_in,
scaler_out=scaler_out)
print('X_test.shape: {0}'.format(X_test.shape))
print('y_test.shape: {0}'.format(y_test.shape))
Let's compute the entire dataset and keep the output data for visualization:
X, y, index, _, _, cols_in_indices, cols_out_indices = prepare_data(
data=df,
lag_data=params['lag_size'],
cols_in=params['features'],
steps_in=params['input_size'],
cols_out=params['features'],
steps_out=params['output_size'],
scaler_in=scaler_in,
scaler_out=scaler_out)
print('X.shape: {0}'.format(X.shape))
print('y.shape: {0}'.format(y.shape))
Let's have a look at the visual representation of the timeseries data:
mwvh.plot_time_series(
title='timeseries visualization',
subtitle='data preparation',
name=('Input - Data Visualization{0}' +
'Training - {1} weeks ({2} samples){0}' +
'Testing - {3} weeks ({4} samples)').format(
'\n',
df_train.shape[0],
X_train.shape[0],
df_test.shape[0],
X_test.shape[0]),
training=df_train,
testing=df_test,
ylabel='feature value (y)',
split=threshold_date)
We'll use RMSE as our loss function to optimize (it is required to be defined as a function that can be compiled by TensorFlow):
def rmse_tf(y_true, y_pred):
return tf.cast(
tf.sqrt(
tf.reduce_mean(
tf.square(
tf.subtract(
y_pred,
y_true)))),
dtype=tf.float32)
First, we define a model-building function. It takes an argument hp from which Keras-Tuner can sample hyperparameters:
def build_model_hp(
hp,
loss_fn,
metrics_fn,
steps_in,
steps_out,
n_features,
n_lstm_layers,
bidirectional_lstm,
n_layers):
# define model
model = km.Sequential()
layer = kl.LSTM(
name='lstm_x',
kernel_regularizer=kr.l2(
l=hp.Float(
name='lstm_x_kernel_regularizer_l2_alpha',
min_value=1e-5,
max_value=1e-3,
default=1e-5,
sampling='log')),
bias_regularizer=kr.l2(
l=hp.Float(
name='lstm_x_bias_regularizer_l2_alpha',
min_value=1e-5,
max_value=1e-3,
default=1e-4,
sampling='log')),
units=hp.Int(
name='lstm_x_units',
min_value=64,
max_value=128,
step=4),
input_shape=(steps_in, n_features),
dropout=hp.Float(
name='lstm_x_dropout',
min_value=0.45,
max_value=0.55,
default=0.5,
step=0.1),
return_sequences=False)
if bidirectional_lstm:
model.add(
kl.Bidirectional(
name='lstm_x_bidirectional',
layer=layer,
merge_mode='ave'))
else:
model.add(layer)
model.add(
kl.RepeatVector(
n=steps_out))
for n_lstm in range(n_lstm_layers):
layer = kl.LSTM(
name='lstm_{0}'.format(n_lstm),
kernel_regularizer=kr.l2(
l=hp.Float(
name='lstm_{0}_kernel_regularizer_l2_alpha'.format(n_lstm),
min_value=1e-5,
max_value=1e-3,
default=1e-5,
sampling='log')),
bias_regularizer=kr.l2(
l=hp.Float(
name='lstm_{0}_bias_regularizer_l2_alpha'.format(n_lstm),
min_value=1e-5,
max_value=1e-3,
default=1e-4,
sampling='log')),
units=hp.Int(
name='lstm_{0}_units'.format(n_lstm),
min_value=64,
max_value=128,
step=4),
dropout=hp.Float(
name='lstm_{0}_dropout'.format(n_lstm),
min_value=0.45,
max_value=0.55,
default=0.5,
step=0.1),
return_sequences=True)
if bidirectional_lstm:
model.add(
kl.Bidirectional(
name='lstm_{0}_bidirectional'.format(n_lstm),
layer=layer,
merge_mode='ave'))
else:
model.add(layer)
dense_dropout = hp.Float(
name='dense_dropout',
min_value=0.45,
max_value=0.55,
default=0.5,
step=0.1)
for n_layer in reversed(range(n_layers)):
layer_size = (1 + n_layer) ** 2
model.add(
kl.TimeDistributed(
name='dense_{0}_timedistributed'.format(n_layer),
layer=kl.Dense(
name='dense_{0}'.format(n_layer),
activation='tanh',
kernel_initializer='glorot_uniform',
bias_initializer='glorot_uniform',
kernel_regularizer=kr.l2(
l=hp.Float(
name='dense_{0}_kernel_regularizer_l2_alpha'.format(n_layer),
min_value=1e-5,
max_value=1e-3,
default=1e-5,
sampling='log')),
bias_regularizer=kr.l2(
l=hp.Float(
name='dense_{0}_bias_regularizer_l2_alpha'.format(n_layer),
min_value=1e-5,
max_value=1e-3,
default=1e-4,
sampling='log')),
units=hp.Int(
name='dense_{0}_units'.format(n_layer),
min_value=8*layer_size,
max_value=16*layer_size,
step=2*layer_size))))
model.add(
kl.Dropout(
name='dense_{0}_dropout'.format(n_layer),
rate=dense_dropout))
model.add(
kl.BatchNormalization(
name='dense_{0}_batchnorm'.format(n_layer)))
model.add(
kl.TimeDistributed(
name='dense_output_timedistributed',
layer=kl.Dense(
name='dense_output',
activation='linear',
kernel_initializer='glorot_uniform',
bias_initializer='glorot_uniform',
kernel_regularizer=kr.l2(
l=hp.Float(
name='dense_output_kernel_regularizer_l2_alpha',
min_value=1e-5,
max_value=1e-3,
default=1e-5,
sampling='log')),
bias_regularizer=kr.l2(
l=hp.Float(
name='dense_output_bias_regularizer_l2_alpha',
min_value=1e-5,
max_value=1e-3,
default=1e-4,
sampling='log')),
units=1)))
model.compile(
loss=loss_fn,
metrics=metrics_fn,
optimizer=ko.Adam(
learning_rate=hp.Float(
name='learning_rate',
min_value=1e-6,
max_value=1e-3,
default=1e-3,
sampling='log')))
return model
It is required to define a partial wrapping the build-function to ensure the signature matches the Keras-Tuner expectations:
build_model = partial(
build_model_hp,
loss_fn=rmse_tf,
metrics_fn=None,
steps_in=params['input_size'],
steps_out=params['output_size'],
n_features=len(params['features']),
n_lstm_layers=1,
bidirectional_lstm=True,
n_layers=1)
We can create an Hyperband tuner to do the hyperparameter search. The main objective is to minimize the validation loss.
tuner = ktt.Hyperband(
build_model,
project_name=params['experiment_name'],
directory='.tuner',
objective='val_loss',
allow_new_entries=True,
tune_new_entries=True,
hyperband_iterations=params['hyperband_iterations'],
max_epochs=params['max_epochs'])
We can start by evaluating a default model, i.e. a model for which all hyperparameters are set to their default value. As we did not specified a default value for all hyperparameters in the build_model_hp function, for such ones their min value will be used instead. To create a default model we just need to pass a fresh kte.hyperparameters.HyperParameters() class instance to the buildfunction.
default_model = tuner.hypermodel.build(
kte.hyperparameters.HyperParameters())
Let's fit this model using the data we loaded earlier:
default_history = default_model.fit(
x=X_train,
y=y_train,
shuffle=True,
batch_size=params['batch_size'],
validation_data=(X_test, y_test),
epochs=params['max_epochs'],
callbacks=[
kc.EarlyStopping(
monitor='val_loss',
patience=params['patience'],
verbose=1,
mode='min',
restore_best_weights=True),
kc.ReduceLROnPlateau(
monitor='val_loss',
factor=1/5,
patience=int(params['patience']/3),
verbose=1,
min_lr=1e-6),
kc.TerminateOnNaN()
],
verbose=2)
We can save this default model and use the Netron application to create a visual representation of it.
default_model.save(
'./.models/{0}_default.h5'.format(
params['experiment_name']))
And here is a visual plot of the model made using the Netron application.
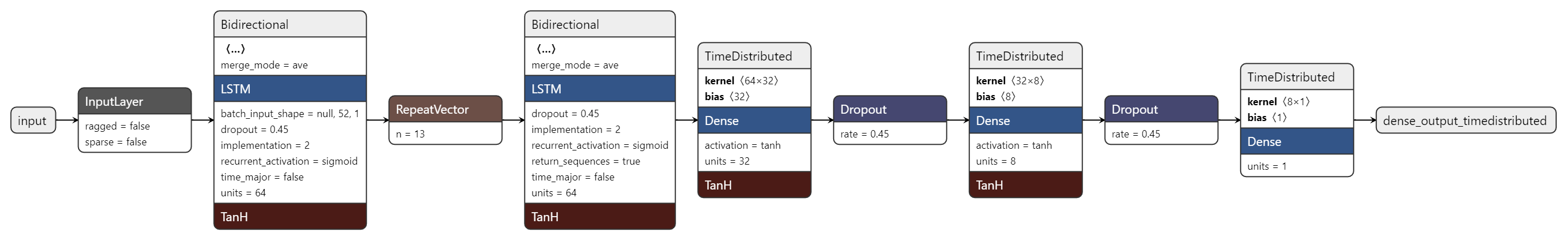
Here is a text version of the model's architecture made with Keras itself.
default_model.summary(
line_length=120)
We can see the optimization process on the chart below.
def plot_model_history(history):
'''
Summarizes the history for all loss functions,
throughout the epochs.
'''
for fn in list(history.history.keys()):
plt.plot(history.history[fn])
plt.title('Model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(list(history.history.keys()), loc='upper right')
plt.show()
plot_model_history(history=default_history)
Let's zoom in and focus on the last n weeks of data.
def get_n_weeks_window(n_weeks):
return dt.timedelta(weeks=n_weeks)
We need a functions to compute the various model scores.
def score(loss_fn, y_true, y_pred):
if loss_fn == 'RMSE':
return skme.mean_squared_error(
y_true=y_true,
y_pred=y_pred,
squared=False)
elif loss_fn == 'MSLE':
return skme.mean_squared_log_error(
y_true=y_true,
y_pred=y_pred)
elif loss_fn == 'MSE':
return skme.mean_squared_error(
y_true=y_true,
y_pred=y_pred,
squared=True)
elif loss_fn == 'R2':
return skme.r2_score(
y_true=y_true,
y_pred=y_pred)
elif loss_fn == 'MAE':
return skme.mean_absolute_error(
y_true=y_true,
y_pred=y_pred)
elif loss_fn == 'MAPE':
return skme.mean_absolute_percentage_error(
y_true=y_true,
y_pred=y_pred)
We can process de predicted values returned by the model. The model sequencialy produces params['output_size'] data points (i.e. the number of weeks of data points predicted). Predictions will enventually overlap, so we can use the mean function to get a good candidate of the current value. Similarly, the min and the max will provide a convenient confidence interval.
def process_predictions(data, X, y, scaler_out, model):
data_pre = data.copy()
scores = []
predictions = []
score_functions = ['RMSE', 'MSE', 'R2', 'MAE', 'MAPE']
lag_data = params['lag_size']
steps_lag = 0
if lag_data is not None and len(lag_data) > 0:
steps_lag = max(lag_data)
y_preds = model.predict(
x=X,
batch_size=params['batch_size'],
verbose=0)
y_true_array = []
y_pred_array = []
for i in range(y.shape[0]):
t_score = {}
y_true = scaler_out.inverse_transform(y[i])
y_pred = scaler_out.inverse_transform(y_preds[i])
y_true_array.append(y_true)
y_pred_array.append(y_pred)
for score_function in score_functions:
t_score[score_function] = score(
score_function,
y_true,
y_pred)
scores.append(t_score)
prediction = [np.nan] * data_pre.shape[0]
for j in range(len(y_true)):
position = steps_lag + i + j + params['input_size']
prediction[position] = y_pred[j][0]
predictions.append(
prediction)
df_scores = pd.DataFrame.from_records(scores)
data_pre['yhat_lower'] = pd.DataFrame(predictions).min().values
data_pre['yhat_upper'] = pd.DataFrame(predictions).max().values
data_pre['yhat'] = pd.DataFrame(predictions).mean().values
data_pre['yhat_last'] = predictions[-1]
data_pre['residual'] = abs(data_pre['yhat'] - data_pre['y'])
pred = {
'df': data_pre,
'scores': df_scores,
'y_true': y_true_array,
'y_pred': y_pred_array,
'predictions': predictions
}
return pred
pred_default = process_predictions(
data=df,
X=X,
y=y,
scaler_out=scaler_out,
model=default_model)
We can see on the chart that the predictions are quite basic but still properly following the local trend as well as the seasonal variations.
mwvh.plot_time_series(
title='Time Series Visualization',
subtitle='default model predictions',
name=('Input - Data Visualization{0}' +
'Training - {1} weeks ({2} samples){0}' +
'Testing - {3} weeks ({4} samples)').format(
'\n',
df_train.shape[0],
X_train.shape[0],
df_test.shape[0],
X_test.shape[0]),
training=df_train,
testing=df_test,
confidence=pred_default['df'],
confidence_label='Confidence interval',
prediction=pred_default['df'],
residual=pred_default['df'],
residual_label='Residual error',
ylabel='feature value (y)',
split=threshold_date,
window_size=get_n_weeks_window(16*params['output_size']))
mwvh.plot_time_series(
title='Time Series Visualization',
subtitle='default model predictions',
name=('Input - Data Visualization{0}' +
'Training - {1} weeks ({2} samples){0}' +
'Testing - {3} weeks ({4} samples)').format(
'\n',
df_train.shape[0],
X_train.shape[0],
df_test.shape[0],
X_test.shape[0]),
testing=df_test,
confidence=pred_default['df'],
confidence_label='Confidence interval',
prediction=pred_default['df'],
prediction_col='yhat',
residual=pred_default['df'],
residual_label='Residual error',
ylabel='feature value (y)',
window_size=get_n_weeks_window(4*params['output_size']))
Keras-Tuner comes with an efficient Hyperband tuner that can search and find the optimized set of parameters. Let's try and find a better model than the default one.
We need this class to clear cell output after each trial:
class ClearTrainingOutput(kc.Callback):
def on_train_end(*args, **kwargs):
ids.clear_output(wait=True)
We can performs a search for best hyperparameters configuration.
tuner.search(
x=X_train,
y=y_train,
shuffle=True,
batch_size=params['batch_size'],
validation_data=(X_test, y_test),
epochs=params['max_epochs'],
callbacks=[
ClearTrainingOutput(),
kc.EarlyStopping(
monitor='val_loss',
patience=params['patience'],
verbose=1,
mode='min',
restore_best_weights=True),
kc.TerminateOnNaN()
],
verbose=2)
We can print out the results summary:
tuner.results_summary(num_trials=1)
And we can retrieve the best hyperparameters configuration:
best_params = tuner.get_best_hyperparameters(num_trials=1)[0]
Which we can use to build the model with the best hyperparameters configuration:
model = tuner.hypermodel.build(best_params)
Let's fit this model using the training data used during the search:
history = model.fit(
x=X_train,
y=y_train,
shuffle=True,
batch_size=params['batch_size'],
validation_data=(X_test, y_test),
epochs=params['max_epochs'],
callbacks=[
kc.EarlyStopping(
monitor='val_loss',
patience=params['patience'],
verbose=1,
mode='min',
restore_best_weights=True),
kc.TerminateOnNaN()
],
verbose=2)
It is a good time to save our model for future use:
model.save(
'./.models/{0}.h5'.format(
params['experiment_name']))
We can see the optimization process on the chart below.
plot_model_history(history=history)
Here are our model scores.
pred_optimized = process_predictions(
data=df,
X=X,
y=y,
scaler_out=scaler_out,
model=model)
We can plot the predictions we've made with our optimized model.
mwvh.plot_time_series(
title='Time Series Visualization',
subtitle='optimized model predictions',
name=('Input - Data Visualization{0}' +
'Training - {1} weeks ({2} samples){0}' +
'Testing - {3} weeks ({4} samples)').format(
'\n',
df_train.shape[0],
X_train.shape[0],
df_test.shape[0],
X_test.shape[0]),
training=df_train,
testing=df_test,
confidence=pred_optimized['df'],
confidence_label='Confidence interval',
prediction=pred_optimized['df'],
residual=pred_optimized['df'],
ylabel='feature value (y)',
split=threshold_date,
window_size=get_n_weeks_window(16*params['output_size']))
This is the last year of data for the optimized model predictions:
mwvh.plot_time_series(
title='Time Series Visualization',
subtitle='optimized model predictions',
name=('Input - Data Visualization{0}' +
'Training - {1} weeks ({2} samples){0}' +
'Testing - {3} weeks ({4} samples)').format(
'\n',
df_train.shape[0],
X_train.shape[0],
df_test.shape[0],
X_test.shape[0]),
testing=df_test,
confidence=pred_optimized['df'],
confidence_label='Confidence interval',
prediction=pred_optimized['df'],
prediction_col='yhat',
residual=pred_optimized['df'],
residual_label='Residual error',
ylabel='feature value (y)',
window_size=get_n_weeks_window(4*params['output_size']))
We can compare both models by zooming in on the last time window, corresponding to the last year of data.
mwvh.plot_time_series(
title='Time Series Visualization',
subtitle='default vs. optimized model predictions',
name=('Input - Data Visualization{0}' +
'Training - {1} weeks ({2} samples){0}' +
'Testing - {3} weeks ({4} samples)').format(
'\n',
df_train.shape[0],
X_train.shape[0],
df_test.shape[0],
X_test.shape[0]),
testing=df_test,
confidence=pred_optimized['df'],
confidence_label='Confidence interval',
prediction=[pred_default['df'], pred_optimized['df']],
prediction_label=['Predicted data (default)', 'Predicted data (optimized)'],
residual=[pred_default['df'], pred_optimized['df']],
residual_label=['Residual error (default)', 'Residual error (optimized)'],
ylabel='feature value (y)',
window_size=get_n_weeks_window(4*params['output_size']))